October 1, 2021 at 11:49 pm | Updated April 11, 2022 at 6:44 am | 6 min read
Precision instruments that take non-destructive measurements of fruit quality parameters are becoming routine throughout global supply chains. Chemometric modeling is a crucial part of this new food science technology. Using applicable chemometric modeling techniqes can increase the accuracy of existing devices by providing better ways of analyzing complex spectral data collected from food. Hence, it is important to stay abreast of the latest developments in the application of these techniques in the food industry.
NIRS & Multiblock Chemometric Modeling
Near Infrared (NIR) is, by far, the most used light band in food science instruments. It interacts with the hydrogen bonds between elements in compounds, making it suitable for qualitative and quantitative analysis of the internal composition of agricultural products.
There are several NIR spectra collected by devices measuring fruit quality. This multivariate data collected by NIR spectroscopy-based devices is analyzed by chemometrics, which combines multivariate statistics, mathematics, and computer science. The models used consider the spectra as one block of variables (X block) that are used to predict quality parameters (Y block).
Subscribe to the Felix instruments Weekly article series.
By submitting this form, you are consenting to receive marketing emails from: . You can revoke your consent to receive emails at any time by using the SafeUnsubscribe® link, found at the bottom of every email. Emails are serviced by Constant Contact
However, at times, spectral data is also collected from compounds’ interaction with visible and short infrared light, etc. This is because different compounds will react with varying wavelengths of light.
Including more than one block of data could improve the width and precision of information available. Multiblock modeling is possible through Artificial Neural Networks (ANN) used in deep machine learning.
Related Article: All About ANN – Artificial Neural Networks and Chemometric Modeling for NIR Spectroscopy
Tinkering with Existing Multiblock Models
Convolutional neural networks (CNNs), a form of ANN, are more powerful than partial least-square regression (PLS), but most of the models so far use only one data block. Two food scientists, Mishra and Passos, wanted to modify CNNs to include multiple blocks by incorporating parallel layers of blocks, with each block getting information from separate sources to deal with unique characteristics of each data type.
This approach has not been attempted before, and the scientists decided to create a model and test it for predicting dry matter in mango.
Constructing the Deep Multiblock Predictive Model
The scientists used a large set of real visible and NIR spectra, which were used as the two parallel blocks of predictive variables. By including the visible spectra, the scientists hoped to get more information on dry matter.
Non-Destructive Measurement of Spectra
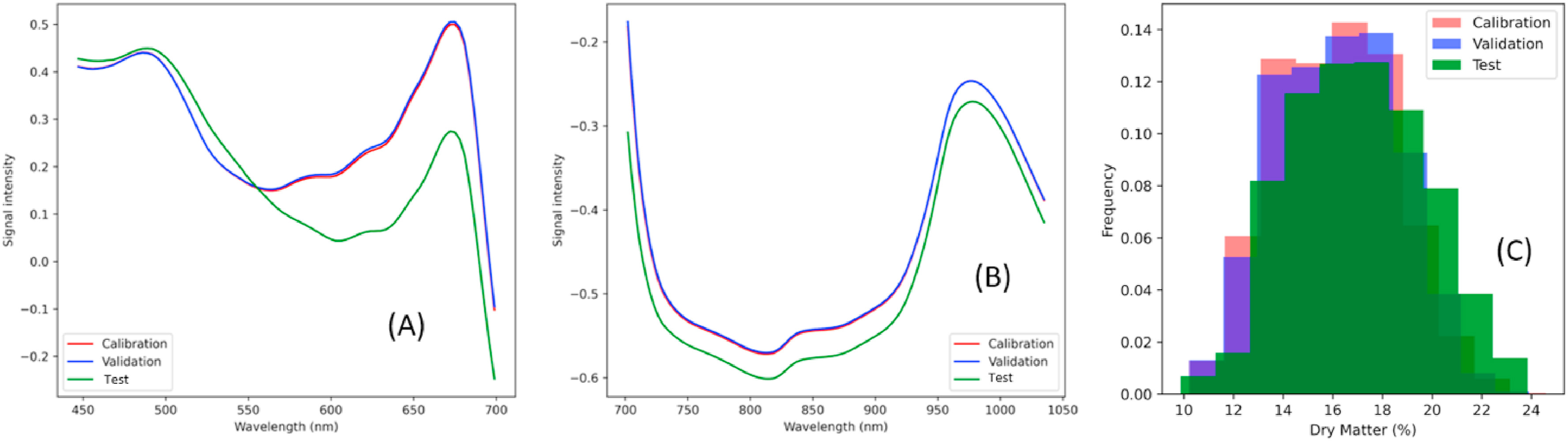
Figure 1: “Mean spectra mango for calibration, validation and test set. (A) Visible, and (B) near-infrared. (C) The histogram of reference dry matter (DM %) for calibration, validation and test set,” Mishra and Passos (2021). (Image credits: doi:10.1016/j.aca.2021.338520)
The spectra are part of an open-source data set. The data was collected by the F-750 Produce Quality Meter, a device that is an industry-standard widely used around the globe in fresh produce supply chains. The NIR spectroscopy device collects information in both visible and NIR spectra to predict dry matter, TSS, and color through non-destructive measurements and is manufactured by Felix Instruments, Applied Food Science.
The scientists used 11,691 spectra in the Vis-NIR range (350–1200 nm). Of this, 10,243 spectra from 2015-2017 were used for training and the 1448 spectra used as the test set were from 2018. The noise was reduced by using data only between 450–1030 nm. Dry matter estimations by oven drying were also made in the laboratory.
To develop the parallel multiblock model, the data was divided into visible spectra (450–697 nm) and NIR spectra (700–1030 nm). See Figure 1 for the differences in the training and test sets of each spectral data. The peaks in the visible spectra are related to the color of the fruit peel, which can vary during ripening from green to yellow and red and are associated with changes in dry matter due to ripening. The peak at 960 nm in the NIR spectra is related to water content, which decreases as dry matter increases.
Steps in Modeling
The training data was prefiltered by Q statistics to remove outliers. The test data was left unfiltered to represent real-life conditions. Two-thirds of the training data was used for calibration and one-third for validation.
The scientists extended the architecture provided by the 1-dimensional convolutional neural network (1D-CNN) to develop the new model.
Convolutional filters (a type of mathematical operation) are key to CNNs and are used to merge data in a block. The scientists reasoned that having parallel blocks allowed them to use separate filters suitable for the data type. For example, NIR data has broad peaks and visual spectroscopy data has sharp peaks. Thus, two mathematical filters of different sizes are needed.
The scientists used two receptive fields where each block of data was analyzed. Each receptive field had one convolutional filter/layer and one stride to produce the feature map. One flattened layer has information pooled from both blocks. There are three fully connected layers operating on this flatten layer, which have 36, 18, and 12 neurons, respectively, to give the output. Figure 2 provides a schematic representation of the steps they used for the modeling.
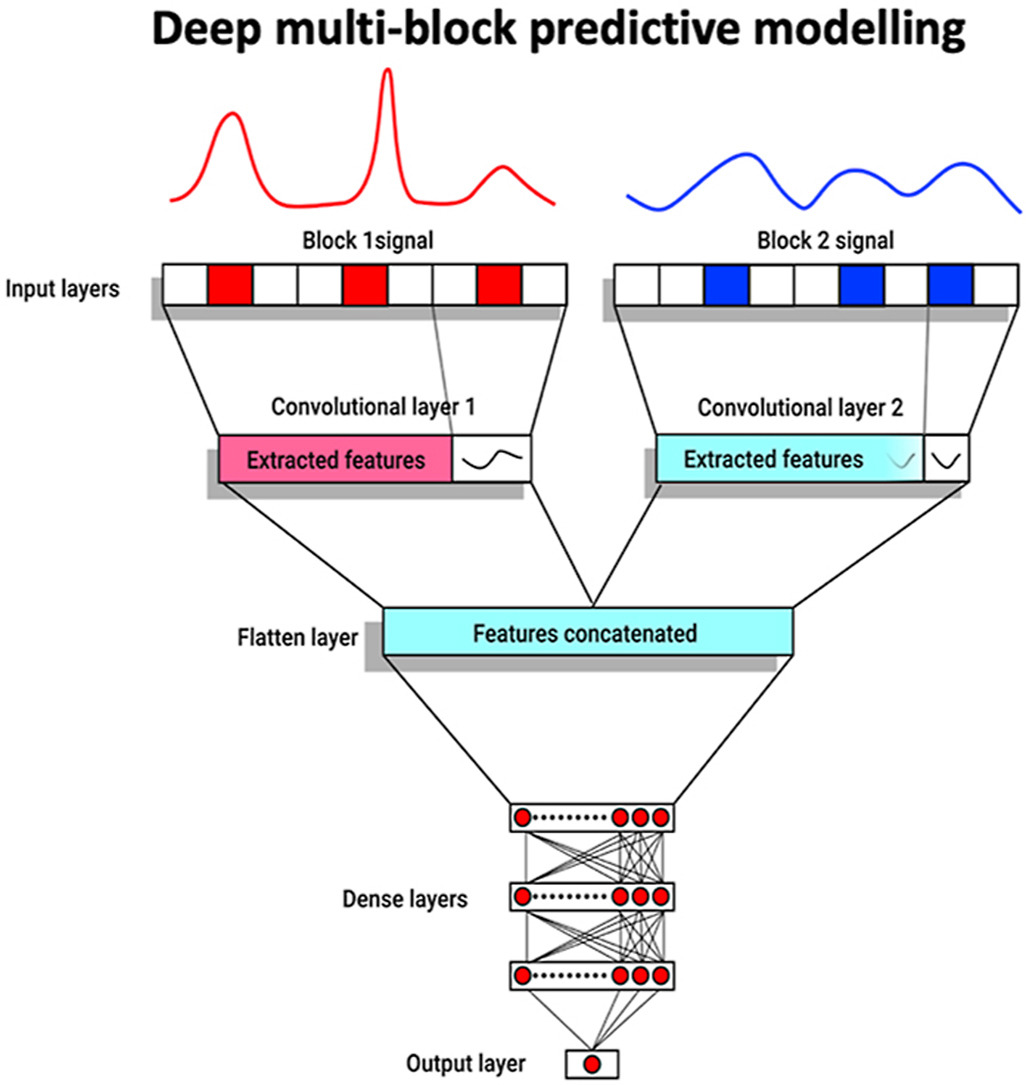
Figure 2: “ A summary of the parallel CNNs architecture used for deep multiblock predictive modeling,” Mishra and Passos (2021). (Image credits: doi:10.1016/j.aca.2021.338520)
The scientists decided to optimize only a few model hyperparameters to provide proof of concept for the new model. They chose to optimize filter sizes for data blocks 1 and 2, the size of the training mini-batch, and the strength of optimization β.
About 1,260 models, with various combinations of the three hyperparameters, were tested by the calibration and validation sets. The models were tested by checking the difference in root mean squared errors (RMSE) of calibration and validation sets. A low difference between the two RMSE was used to choose the best optimal β, batch size, and filter size.
The performance of the new deep parallel multiblock model was compared with the following:
- a traditional single block CNN
- a multiblock chemometric approach: the sequentially orthogonalized partial least-squares (SO-PLS) regression
- previous best models created by other scientists, using only the NIR part of the same mango open-source data
Comparing Chemometric Models
When the scientists compared the various models, they found that the deep multiblock model outperformed all the other models it was tested against. Among all models tested, it had the lowest RMSE of 0.818%.
The scientists found the deep multiblock model was better than the models created by other researchers in previous studies with the same NIR data set, where the RMSE = 0.84%. This illustrates the usefulness of making data public, so that it can be used for different experiments. By using the same data, it is possible to accurately identify the strengths and weaknesses of models. Dissimilar conditions and data are a bane in comparing scientific results, and this current study has solved this problem elegantly.
The single block CNN performed better than the multiblock chemometric approach. The latter uses a linear model that does not take into account the nonlinearity in the spectral data, whereas even a single block CNN will do so.
The deep multiblock CNN was most influenced by optimal β. So, this hyperparameter was first identified, and then later models with the best filter width and batch size were chosen. The scientists found the least difference in the RMSE of the calibration and validation sets occurred at β= 0.003 and batch size 32. The best filter width was 5 for the visible range and 30 for NIR.
The study shows that the multiblock method is superior to the single block CNN. The scientists also identified areas of modeling which they think require further attention, such as optimization. They were, however, able to prove that different filters for separate data sets can be a major advantage. The number of blocks is not an issue, and future models can have many, even if computation time increases. The scientists are confident that the parallel block model can also be applied to 2-D and 3-D data.
Improving Model Precision
By segregating the existing data and extending models already in use, it is possible to make NIR spectroscopy devices even more accurate while retaining existing hardware. Continued development of chemometric techniques using industry-standard technology like the F-750 Produce Quality Meter improves the precision that can be achieved in food science research and food supply chains.
—
Vijayalaxmi Kinhal
Science Writer, CID Bio-Science
Ph.D. Ecology and Environmental Science, B.Sc Agriculture
Sources
Dertat, A. ( 2017, Nov 8). Applied Deep Learning – Part 4: Convolutional Neural Networks. Retrieved from https://towardsdatascience.com/applied-deep-learning-part-4-convolutional-neural-networks-584bc134c1e2
Puneet Mishra, & Passos, D. (2021). Deep multiblock predictive modelling using parallel input convolutional neural networks. Analytica Chimica Acta, 1163, 338520. doi:10.1016/j.aca.2021.338520
Related Products
- F-751 Grape Quality Meter
- Custom Model Building
- F-901 AccuRipe & AccuStore
- F-751 Melon Quality Meter
- F-751 Kiwifruit Quality Meter
- F-750 Produce Quality Meter
- F-751 Avocado Quality Meter
- F-751 Mango Quality Meter
- F-900 Portable Ethylene Analyzer
- F-950 Three Gas Analyzer
- F-920 Check It! Gas Analyzer
- F-960 Ripen It! Gas Analyzer
- F-940 Store It! Gas Analyzer
Most Popular Articles
- Spectrophotometry in 2023
- The Importance of Food Quality Testing
- NIR Applications in Agriculture – Everything…
- The 5 Most Important Parameters in Produce Quality Control
- Melon Fruit: Quality, Production & Physiology
- Guide to Fresh Fruit Quality Control
- Liquid Spectrophotometry & Food Industry Applications
- Ethylene (C2H4) – Ripening, Crops & Agriculture
- Fruit Respiration Impact on Fruit Quality
- Active Packaging: What it is and why it’s important






